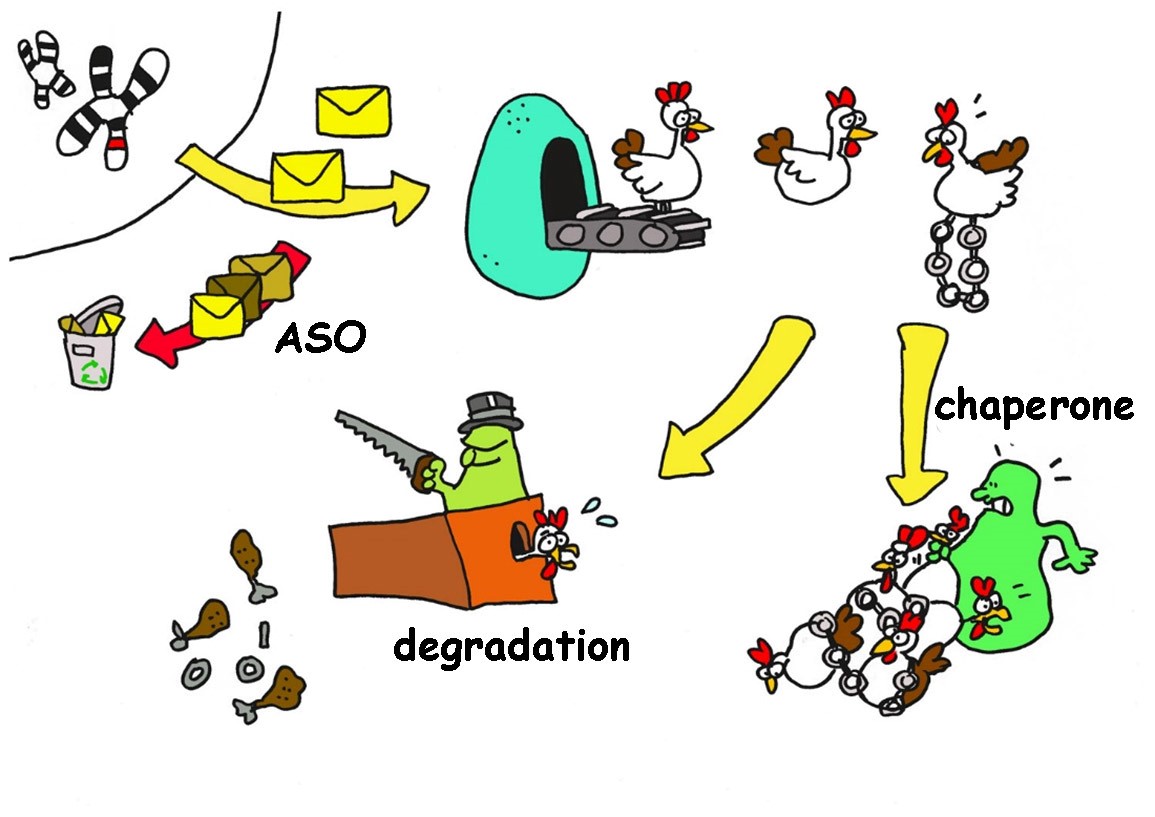
REITS GROUP – IMPROVING PROTEIN DEGRADATION IN HUNTINGTON’S DISEASE
Huntington’s disease (HD) is hallmarked by the accumulation and aggregation of mutant huntingtin protein (mHTT) fragments. Our goal is to improve recognition and degradation of mHTT by the ubiquitin-proteasome system.
Contact: E.A. Reits (e.a.reits@amc.uva.nl)
Huntington’s Disease (HD) is an dominant inherited neurodegenerative disorder with the combined symptoms of Alzheimer, Parkinson and ALS. HD is caused by a mutation in the gene encoding the huntington protein. Due to the expansion of the CAG repeat in the gene the encoded huntington protein had an extended repeat of glutamine amino acids (polyQ) in the protein. Due to the repeat expansion the mutant Htt protein (mHtt) aggregates in neuronal cells, leading to their dysfunction, with consequences for memory and movement and psychiatric problems. Following the first symptoms the patient will live on average 15-17 years.
Improving the degradation of these mHtt fragments prior to aggregation would be a therapeutic strategy for this devastating disease for which there is no cure.
The main protein degradation machinery in cells is the Ubiquitin-Proteasome System (UPS) which is present in both the cytoplasm and nucleus. It degrades both short-lived, misfolded proteins and long-lived proteins that are mainly targeted for degradation via ubiquitination. The role of the UPS in HD is however controversial since UPS impairment has been observed in various HD models, which could be due to sequestration of proteasomes into aggregates (inclusion bodies, IB) that are initiated by mHTT. In addition, proteasomes may even be unable to cleave within the polyglutamine (polyQ) expansion in mHTT that is caused by the CAG repeat expansion in the mutated gene. Proteasomes may become clogged by these fragments or release polyQ peptides when polyQ-expanded proteins are inefficiently degraded.
Our research group have addressed various research questions with relation to the UPS and its role in the degradation of mHtt.
1. Proteasomes are able to degrade mHtt entirely
Next we examined whether proteasomes are indeed unable to degrade polyQ-expanded htt fragments in living cells, using modified htt fragments that were exclusively targeted to proteasomes (and not to autophagosomes). Interestingly, these htt fragments were efficiently and completely degraded, which was also observed in vitro using purified htt fragments. The degradation may however occur in various steps, as initially the flanking amino acids were degraded, while the remaining polyQ fragment was degraded in time.
Published in JBC 2013: Expanded Polyglutamine-containing N-terminal Huntingtin Fragments Are Entirely Degraded by Mammalian Proteasomes.

2. Chaperones assist in the degradation of mHtt
We examined whether there are chaperones that could prevent the aggregation of polyQ peptides. We found that the small DnaJB6 and DnaJB8 chaperones prevent polyQ peptide aggregation, thereby enabling their degradation. Intriguingly, these chaperones act directly on polyQ peptides but only indirectly on polyQ-expanded htt fragments, supporting earlier in vitro data using purified proteasomes that polyQ peptides may initiate aggregation.
Published in JBC 2013: The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides.

3. Proteasomes are dynamically recruited into mHtt aggregates and remain active
To examine the reported sequestration of proteasomes into polyQ-expanded htt aggregates in more detail, we performed fluorescent pulse-chase experiments in living cells to study the dynamics of proteasomes over longer time spans. In contrast to earlier reports, we observed that proteasomes are reversibly recruited into aggregates, and in addition are still active and accessible for substrates. This would explain the absence of UPS impairment in HD models when aggregates are observed, and indicates that proteasomes are actively recruited and remain active.
Published in FEBS 2014: Dynamic recruitment of active proteasomes into polyglutamine initiated inclusion bodies.

4. Ubiquitination of mHtt
Ubiquitination of proteins can be a signal for degradation and relies on its mono- and polymeric isoforms attached to protein substrates. Studying the ubiquitination pattern of aggregated Htt fragments offers an important possibility to understand Htt degradation and aggregation processes within the cell. For the identification of aggregated Htt and its ubiquitinated species, solubilization of the cellular aggregates is mandatory. We generated methods to identify post-translational modifications such as ubiquitination of aggregated mutant Htt, and showed that mHtt is poly-ubiquitinated when it is aggregated.
Currently we are examining how soluble mHtt and wtHtt is ubiquitinated in different brain areas, which (de)ubiquitinating enzymes are involved, and how to improve mHtt ubiquitination and degradation.
Published in Front Mol Neurosci: Detection of ubiquitinated huntingtin species in intracellular aggregates.

5. Differences in mHtt aggregation between different cell types
While mutant huntingtin protein (mHTT) is ubiquitously expressed the effects of mHtt aggregates have been mainly studied in neurons. When we examined the number of mHTT inclusions in both neurons and various types of glia cells in the brain of different HD mouse models and HD patient brains, we observed that half of the population of neurons contained nuclear inclusions at the disease end stage, whereas the proportion of astrocytes and oligodendrocytes having a nuclear inclusion was much lower, while microglia hardly showed any nuclear inclusions. This indicates that different cell types can handle mHtt differently and thus with better efficiency.
Published in Glia 2017: Frequency of nuclear mutant huntingtin inclusion formation in neurons and glia is cell-type-specific.

6. Dynamic (de)ubiquitination events in mHtt aggregates
We also showed dynamic recruitment of ubiquitin and ubiquitin (de)-conjugating activity at mHtt initiated IBs. Using synthesized TAMRA-labeled ubiquitin moieties, we observed that intracellular TAMRA-ubiquitin is dynamic at mHtt IBs and is incorporated into poly-ubiquitin chains of intracellular substrates such as mHtt. mHtt IBs recruit catalytically active enzymes involved in (de)-ubiquitination processes, which we visualised using novel activity-based probes. In contrast to TAMRA-Ub, the larger GFP-ubiquitin reporter used in earlier studies becomes irreversibly sequestered, suggesting a methodical disadvantage of GFP-tagged ubiquitin.
Published in Scientific Reports 2018: Dynamic recruitment of ubiquitin to mutant huntingtin inclusion bodies.

Current research
Since the UPS appear to work properly in HD, with proteasomes and ubiquitin being dynamically recruited to mHtt aggregates, and the proteasome able to degrade mHtt when it is marked for destruction by ubiquitin, we are currently addressing the following questions:
- Can proteasome activity be manipulated to improve mHtt degradation? We observe differences in the degradation of mHtt by different proteasome complexes in vitro, and also observe changes in proteasome complex formation during HD progression.
- How is mHtt ubiquitinated, and which (de)ubiquitinating enzymes are involved? Wild-type Htt (wtHtt) and mHtt seem differently ubiquitinated and with different efficiencies, and identifying involved enzymes may allow us to manipulate and improve mHtt ubiquitination and degradation
- Screening with compounds and RNA knockdown to improve degradation of (untagged) mHtt fragments. Using neuronal cell-based assays expressing untagged Htt fragments we aim to identify involved (de)ubiquitinating enzymes and modify their activities to improve mHtt degradation.
AMC-VUmc Research Institute: Amsterdam Neuroscience
The research group is part of the Amsterdam Neuroscience which includes the AMC, VUmc, VU, UvA and Amsterdam Neuroscience Institute researchers and clinicians.
Funding
The described research is currently funded by the Campagneteam Huntington and CHDI, and previously by NWO (VIDI), Hersenstichting, Prinses Beatrix Fondsn, de Vereniging van Huntington, an AMC PhD scholarship, and earlier by a grant from the KWF, HDF and a NWO-VENI-grant.

- Dynamic recruitment of ubiquitin to mutant huntingtin inclusion bodies. Juenemann K, Jansen AHP, van Riel L, Merkx R, Mulder MPC, An H, Statsyuk A, Kirstein J, Ovaa H, Reits EA. Sci Rep. 2018 Jan 23;8(1):1405 www.ncbi.nlm.nih.gov/pubmed/29362455
- Visualization of prion-like transfer in Huntington’s disease models. Jansen AH, Batenburg KL, Pecho-Vrieseling E, Reits EA. Biochim Biophys Acta. 2017 Mar;1863(3):793-800 www.ncbi.nlm.nih.gov/pubmed/28040507
- Frequency of nuclear mutant huntingtin inclusion formation in neurons and glia is cell-type-specific. Jansen AH, van Hal M, Op den Kelder IC, Meier RT, de Ruiter AA, Schut MH, Smith DL, Grit C, Brouwer N, Kamphuis W, Boddeke HW, den Dunnen WF, van Roon WM, Bates GP, Hol EM, Reits EA. Glia. 2017 Jan;65(1):50-61 www.ncbi.nlm.nih.gov/pubmed/27615381
- Tripeptidyl Peptidase II Mediates Levels of Nuclear Phosphorylated ERK1 and ERK2. Wiemhoefer A, Stargardt A, van der Linden WA, Renner MC, van Kesteren RE, Stap J, Raspe MA, Tomkinson B, Kessels HW, Ovaa H, Overkleeft HS, Florea B, Reits EA. Mol Cell Proteomics. 2015 Aug;14(8):2177-93 www.ncbi.nlm.nih.gov/pubmed/26041847
- Detection of ubiquitinated huntingtin species in intracellular aggregates. Juenemann K, Wiemhoefer A, Reits EA. Front Mol Neurosci. 2015 Jan 28;8:1 www.ncbi.nlm.nih.gov/pubmed/25674046
- The ubiquitin proteasome system in glia and its role in neurodegenerative diseases. Jansen AH, Reits EA, Hol EM. Front Mol Neurosci. 2014 Aug 8;7:73 www.ncbi.nlm.nih.gov/pubmed/25152710
- Dynamic recruitment of active proteasomes into polyglutamine initiated inclusion bodies. Schipper-Krom S, Juenemann K, Jansen AH, Wiemhoefer A, van den Nieuwendijk R, Smith DL, Hink MA, Bates GP, Overkleeft H, Ovaa H, Reits E . FEBS Lett. 2014. www.ncbi.nlm.nih.gov/pubmed/24291262
- Expanded polyglutamine-containing N-terminal huntingtin fragments are entirely degraded by mammalian proteasomes. Juenemann K, Schipper-Krom S, Wiemhoefer A, Kloss A, Sanz Sanz A and Reits EA. J Biol Chem. 2013 www.ncbi.nlm.nih.gov/pubmed/23908352
- The DNAJB6 and DNAJB8 chaperones prevent intracellular aggregation of polyglutamine peptides. Gillis J, Schipper-Krom S, Juenemann K, Gruber A, Coolen S, van Nieuwendijk R, van Veen H, Overkleeft H, Goedhart J, Kampinga HH, Reits EA. J Biol Chem. 2013. www.ncbi.nlm.nih.gov/pubmed/23612975
- Reduced amyloid-β degradation in early Alzheimer’s disease but not in the APPswePS1dE9 and 3xTg-AD mouse models. Stargardt A, Gillis J, Kamphuis W, Wiemhoefer A, Kooijman L, Raspe M, Benckhuijsen W, Drijfhout JW, M Hol E, Reits E. Aging Cell. 2013 www.ncbi.nlm.nih.gov/pubmed/23534431
- The Ubiquitin-Proteasome System in Huntington’s Disease: Are Proteasomes Impaired, Initiators of Disease, or Coming to the Rescue? Schipper-Krom S, Juenemann K, Reits EA. Biochem Res Int. 2012;2012:837015 www.ncbi.nlm.nih.gov/pubmed/23050151
- Aminopeptidase-resistant peptides are targeted to lysosomes and subsequently degraded. Gillis JM, Benckhuijsen W, van Veen H, Sanz AS, Drijfhout JW, Reits EA. Traffic. 2011 www.ncbi.nlm.nih.gov/pubmed/21883763
- Mimicking proteasomal release of polyglutamine peptides initiates aggregation and toxicity. Raspe M, Gillis J, Krol H, Krom S, Bosch K, van Veen H, Reits E. J Cell Sci. 2009 www.ncbi.nlm.nih.gov/pubmed/19690053
- PKC gamma mutations in spinocerebellar ataxia type 14 affect C1 domain accessibility and kinase activity leading to aberrant MAPK signaling. Verbeek DS, Goedhart J, Bruinsma L, Sinke RJ, Reits EA.J Cell Sci. 2008 Jul 15;121(Pt 14):2339-49 www.ncbi.nlm.nih.gov/pubmed/18577575
- Polyglutamine expansion accelerates the dynamics of ataxin-1 and does not result in aggregate formation. Krol HA, Krawczyk PM, Bosch KS, Aten JA, Hol EM, Reits EA. PLoS One. 2008 Jan 30;3(1):e1503 www.ncbi.nlm.nih.gov/pubmed/18231590
- Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. Reits EA, Hodge JW, Herberts CA, Groothuis TA, Chakraborty M, Wansley EK, Camphausen K, Luiten RM, de Ru AH, Neijssen J, Griekspoor A, Mesman E, Verreck FA, Spits H, Schlom J, van Veelen P, Neefjes JJ. J Exp Med. 2006 May 15;203(5):1259-71 www.ncbi.nlm.nih.gov/pubmed/16636135
- Monitoring the distribution and dynamics of proteasomes in living cells. Groothuis TA, Reits EA. Methods Enzymol. 2005;399:549-63. www.ncbi.nlm.nih.gov/pubmed/16338381
- A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Reits E, Neijssen J, Herberts C, Benckhuijsen W, Janssen L, Drijfhout JW, Neefjes J. Immunity. 2004 www.ncbi.nlm.nih.gov/pubmed/15084277
- Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Immunity. 2003 Jan;18(1):97-108. www.ncbi.nlm.nih.gov/pubmed/12530979
- From fixed to FRAP: measuring protein mobility and activity in living cells. Reits EA, Neefjes JJ. Nat Cell Biol. 2001 www.ncbi.nlm.nih.gov/pubmed/11389456
- The major substrates for TAP in vivo are derived from newly synthesized proteins. Reits EA, Vos JC, Grommé M, Neefjes J. Nature. 2000 www.ncbi.nlm.nih.gov/pubmed/10783892
- Dynamics of proteasome distribution in living cells. Reits EA, Benham AM, Plougastel B, Neefjes J, Trowsdale J. EMBO J. 1997 www.ncbi.nlm.nih.gov/pubmed/9321388
The core facility Cellular Imaging at the AMC harbors all the advanced fluorescence microscopy, electron microscopy and flow cytometry in one facility and is currently facilitating almost 600 active users. The facility is headed by Eric Reits and has a staff of 10. Techniques include:
Advanced fluorescence microscopy
- Confocal microscopy
- Superresolution microscopy
- FLIM imaging
- Automated live cell microscopy
- Lightsheet microscopy
- Inverted and upright fluorescence microscopy
Electron microscopy
- Transmission EM
- Scanning EM
- Correlative Light and Electron Microscopy (CLEM)
- Immuno EM
- Cryo EM
While Alzheimer and Parkinson are in general not inherited diseases and occur ‘spontaneously’ while aging, Huntington’s Disease (HD) is a dominant inherited disease and children of a parent with the HD-related mutation (gene-carrier) have 50% chance to inherit the disease. Due to the symptoms, the absence of a cure but also the direct consequences for someone’s live (e.g. for relations, choices in life like studying or not, mortgage, insurances) only 10-15% of these so-called risk-carriers want to know whether they are in fact gene-carrier. As a result of this taboo, the mutated gene is silently passed on within families. The taboo also limits the willingness to raise awareness by public campaigns, and therefore also to raise funding for research.
In 2016 the Campagneteam Huntington was founded by many volunteers from families with Huntington’s Disease (HD), with the goal to raise awareness for the disease and to raise funds for research aiming for a cure for HD. There were various reasons including:
- Funding for HD research evaporated when the Prinses Beatrix Foundation (movement disorders) decided to focus on muscle diseases only, which excluded diseases like HD and ALS that affect movement
- In contrast to ALS, Parkinson and Alzheimer there was no awareness campaign for HD, partly due to the huge taboo on the disease. Being a 100% genetic disorder with 50% chance of inheritance, with various symptoms ranging from psychiatric problems and depression to chorea, and the absence of any treatment, most persons at risk do not want a genetic test. One can test at the age of 18, but many prefer not to know what may lay ahead and will affect relations, career and insurances.
- There were various promising research lines by different Dutch HD groups aiming to reduce mutant huntingtin levels by either lowering the synthesis of the protein, prevent aggregation of the mutant protein, or improve its degradation. These research lines were jeopardized when funding dropped.
With the free help of the advertising agency Artica and marketing professionals like Roger Verdurmen the volunteers of the Campagneteam Huntington worked on a campaign including a television commercial, street commercials and flyers, and a website. Designing a slogan to describe the disease was not easy, as how to visualize a disease with the combined symptoms of ALS, Alzheimer and Parkinson, and the huge taboo in most families affected by the disease? It turned out that HD cannot be visualized but can better be described:
A disease so awful that no-one talks about it
The word ‘doodgezwegen’ stands for being dead silence, not speaking about it at all, being a taboo.
Foundation Campagneteam Huntington
The foundation Campagneteam Huntington was raised on March 22 2016 in order to acquire an ANBI bank account (Public benefit organization, allowing people and companies to deduct their gift from their taxable income). Together with the HD researchers in the Netherlands (Huntingtonresearch.nl) the Campagneteam Huntington aims to finance (fundamental) research needed to find a cure for HD and has the following objectives:
- Organise fundraising activities for research aiming for a cure for HD.
- Organising a national campaign to create awareness for HD by the general public.
- Finance research projects that are selected and approved by the independent Scientific Advisory Board
The foundation explicitly does not aim to make a profit, and is run by volunteers including the board. All raised funds will entirely be spent on funding research projects that are chosen by the independent Scientific Advisory Board comprised of neuroscientists from related research fields that are not working on HD themselves. The board is formed by Melanie Kroezen (chair), Marijke Heefer (secretary), Ivo Braakhuis (treasurer) and Eric Reits (as chair of the Dutch HuntingtonResearch network).
Campaign and activities
The campaign was launched in May 2016, and raised much attention in the national and local media. Hundreds of fundraising activities have been initiated by even more volunteers throughout the country, and these activities are listed on the website. Besides these heart-warming activities also large donations were made by families and foundations, including gifts of 40.000 to 250.000 euro’s to finance complete research lines. Within 2 years more than 2 million euros were donated, which enabled the financing of 6 research lines already and an open call in 2018 for more scientists to work on a cure for HD.
Scientific developments
For decades most research was aiming for treating symptoms, and studying the various consequences of mutant huntington protein expression in cells (that affects many cellular processes such as vesicle transport and mitochondrial functioning). Various potential compounds were tested in trials but none were effective, or can only be used to treat particular symptoms (e.g. depression). However, more recent research focuses on the cause of disease: the mutant huntington protein itself. The central aim is to decrease synthesis of the mutant protein, prevent aggregation of the mutant protein, and/or improve degradation of the mutant protein.
There are various research lines thinkable (and ongoing) that aim for a cure for HD, including:
- Inhibition of synthesis of the mutant huntington protein: making use of so-called interference or antisense RNA the signal between de corresponding mutated gene and the protein synthesis machinery is intercepted. There are various strategies that can be exploited in HD, and the most well-known is the IONIS phase I clinical trial that is now continued by Roche.
- Preventing aggregation of the mutant huntington protein. Or cells express so-called chaperones that assist in protein folding, and the prevention of protein aggregation. Some chaperones were shown to prevent aggregation of the mutant huntington protein, and a therapeutic strategy would be to induce these chaperones in the brain. It was recently published that overexpression of one of these chaperones delayed onset of HD in animal models.
- Improving recognition and degradation of mutant huntington proteins. All proteins are degraded sooner or later by a protease, an enzyme that recycles proteins. In order to be recognized for degradation these proteins need to be labelled by a marker, and preliminary data indicates that due to the mutation the huntington protein is inefficiently labelled, and therefor hardly recognized for degradation. Identifying involved enzymes and stimulating proper labelling and degradation would be a therapeutic strategy to accelerate mutant huntington protein degradation. Interestingly, while the mutant huntington protein is synthesized in most cells, only particular cells are affected, while others seem more efficient in degrading mutant huntington proteins. This could be due to differences in the levels of involved enzymes, or chaperones.
- Screening with existing medicines/compounds. Using large libraries of existing compounds (used/developed for other disease studies) their effect on reducing mutant huntington protein levels can be tested. Effective compounds must be verified, and their mechanism explored (how, via what enzyme, specificity).
Note that all these research lines aim to reduce the level of mutant huntington protein, which is the direct cause of disease.